Ciwon Daji Epigenetics
|
specialty (en) | |
| Bayanai | |
| Ƙaramin ɓangare na |
academic discipline (en) |
| Bangare na |
epigenetics (en) |
| Is the study of (en) |
gene expression (en) |
Ciwon daji epigenetics shine nazarin gyare-gyaren epigenetic zuwa DNA na kwayoyin cutar kansa wanda ba ya haɗa da canji a cikin jerin nucleotide, amma a maimakon haka ya haɗa da canji a yadda aka bayyana lambar kwayoyin halitta. Hanyoyin Epigenetic suna da mahimmanci don kula da jerin al'ada na takamaiman maganganun kwayoyin halitta na nama kuma suna da mahimmanci don ci gaba na al'ada. [1] Wataƙila suna da mahimmanci, ko ma mafi mahimmanci, fiye da maye gurbin kwayoyin halitta a cikin canjin tantanin halitta zuwa kansa. Rikicin hanyoyin epigenetic a cikin cututtukan daji, na iya haifar da asarar bayyanar kwayoyin halittar da ke faruwa kusan sau 10 akai-akai ta hanyar yin shiru na rubutu (wanda ya haifar da haɓakar epigenetic hypermethylation na tsibiran CpG ) fiye da maye gurbi. Kamar yadda Vogelstein et al. nuna, a cikin ciwon daji na launin fata yawanci ana samun kusan maye gurbin direba 3 zuwa 6 da 33 zuwa 66 hitchhiker ko maye gurbin fasinja.[2] Duk da haka, a cikin ciwace-ciwacen hanji idan aka kwatanta da kusa da mucosa na al'ada na al'ada, akwai kimanin 600 zuwa 800 tsibiran methylated CpG a cikin masu inganta kwayoyin halitta a cikin ciwace-ciwacen ƙwayoyi yayin da waɗannan tsibiran CpG ba su da methylated a cikin mucosa kusa.[3][4][5] Yin amfani da sauye-sauye na epigenetic yana kuma riƙe da babban alƙawari don rigakafin ciwon daji, ganowa, da jiyya.[6][7] A cikin nau'ikan ciwon daji daban-daban, nau'ikan nau'ikan nau'ikan epigenetic na iya rikicewa, kamar yin shiru na ƙwayoyin cuta masu hana ƙari da kunna oncogenes ta hanyar canza tsarin methylation na tsibirin CpG, gyare-gyaren histone, da dysregulation na sunadaran ɗaurin DNA. Ana amfani da magunguna da yawa waɗanda ke da tasirin epigenetic yanzu a yawancin waɗannan cututtuka.


Makanikai[gyara sashe | gyara masomin]
DNA methylation[gyara sashe | gyara masomin]
A cikin sel somatic, tsarin DNA methylation gabaɗaya ana watsa su zuwa sel 'ya'ya masu aminci.[8] Yawanci, wannan methylation yana faruwa ne kawai a cytosines waɗanda suke 5' zuwa guanosine a cikin CpG dinucleotides na eukaryotes mafi girma.[9] Koyaya, methylation DNA na epigenetic ya bambanta tsakanin ƙwayoyin al'ada da ƙwayoyin ƙari a cikin mutane. Bayanan martaba na "al'ada" CpG methylation galibi ana jujjuya su a cikin sel waɗanda suka zama tumorigenic. A cikin sel na al'ada, tsibiran CpG da ke gaba da masu tallata kwayoyin halitta gabaɗaya ba su da ƙarfi, kuma suna yin aiki ta hanyar rubutawa, yayin da sauran CpG dinucleotides ɗin kowane mutum a cikin kwayar halitta yakan zama methylated. Koyaya, a cikin ƙwayoyin cutar kansa, tsibiran CpG waɗanda ke gaba da masu hana ƙwayoyin cuta ƙwayoyin cuta galibi suna hypermethylated, yayin da CpG methylation na yankuna masu haɓaka oncogene da jerin maimaita parasitic galibi suna raguwa.[10]
Hypermethylation na yankuna masu hana ƙwayar ƙwayar cuta na iya haifar da yin shuru ga waɗannan ƙwayoyin. Irin wannan maye gurbi na epigenetic yana ba da damar sel suyi girma da kuma haifuwa ba tare da kulawa ba, yana haifar da tumorigenesis. Ƙarin ƙungiyoyin methyl zuwa cytosines yana haifar da DNA don yin murɗawa kusa da sunadaran histone, wanda ke haifar da DNA wanda ba zai iya jujjuya rubutu ba (DNA da aka yi shiru). Kwayoyin halittar da aka saba da su don yin shuru a rubuce saboda masu haɓaka hypermethylation sun haɗa da: Cyclin-dependent kinase inhibitor p16, mai hana sake zagayowar tantanin halitta; MGMT, kwayar gyaran DNA ; APC, mai kula da zagayawa; MLH1, kwayar halittar DNA-gyaran; da BRCA1, wani DNA-gyaran kwayoyin halitta. Lallai, ƙwayoyin kansa na iya zama masu sha'awar yin shuru na rubutu, saboda mai haɓaka hypermethylation, na wasu mahimman ƙwayoyin cuta masu hana ƙari, wani tsari da aka sani da jarabar epigenetic.[11]
Hypomethylation na CpG dinucleotides a cikin sauran sassan kwayoyin halitta yana haifar da rashin zaman lafiyar chromosome saboda hanyoyin kamar asarar bugawa da sake kunna abubuwa masu iya canzawa . Rashin buguwar kwayar halitta mai girma kamar insulin (IGF2) yana ƙara haɗarin ciwon daji na launin fata kuma yana da alaƙa da ciwon Beckwith-Wiedemann wanda ke ƙara haɗarin ciwon daji ga jarirai. A cikin sel lafiya, CpG dinucleotides na ƙananan yawa ana samun su a cikin coding da kuma wuraren da ba sa coding intergenic. Bayanin wasu jerin maimaitawa da sake haduwar meiotic a centromeres ana murƙushe su ta hanyar methylation[12]
Dukkanin kwayoyin halitta na kwayar cutar kansa ya ƙunshi ƙarancin methylcytosine fiye da kwayar halitta mai lafiya. A gaskiya ma, kwayoyin cutar kansa suna da 20-50% kasa da methylation a kowane CpG dinucleotides a fadin kwayoyin halitta. Tsibirin CpG da aka samu a yankuna masu tallatawa galibi ana kiyaye su daga DNA methylation. A cikin ƙwayoyin kansa tsibiran CpG tsibiran suna hypomethylated Yankunan da ke gefen tsibiran CpG da ake kira tsibirin tsibirin CpG sune inda yawancin methylation na DNA ke faruwa a cikin mahallin CpG dinucleotide. Kwayoyin ciwon daji suna da methylated ta hanyar da ba ta dace ba a tsibirin CpG. A cikin kwayoyin cutar kansa, hypermethylation a cikin tsibirin CpG tsibirin suna motsawa zuwa tsibiran CpG, ko hypomethylation na tsibiran CpG suna motsawa zuwa tsibirin CpG tsibiri suna kawar da iyakokin epigenetic tsakanin waɗannan abubuwan halitta.[13] A cikin kwayoyin cutar kansa "hypomethylation na duniya" saboda rushewa a cikin DNA methyltransferases (DNMTs) na iya haɓaka haɓakar mitotic da sake tsarawa na chromosome, a ƙarshe yana haifar da aneuploidy lokacin da chromosomes suka kasa rabuwa da kyau yayin mitosis .
CpG tsibirin methylation yana da mahimmanci a cikin tsarin maganganun kwayoyin halitta, duk da haka cytosine methylation na iya haifar da kai tsaye zuwa lalata maye gurbi da yanayin salon salula na farko. Methylated cytosines suna sa hydrolysis na rukunin aminin da jujjuyawar kai tsaye zuwa thymine ya fi dacewa. Suna iya haifar da ɗaukar ma'aikata na furotin chromatin . Cytosine methylations yana canza adadin hasken UV na tushen nucleotide, haifar da dimers pyrimidine . Lokacin da maye gurbi ya haifar da asarar heterozygosity a rukunin yanar gizon masu hana ƙari ya kasan ce shi ne waɗannan kwayoyin halitta na iya zama marasa aiki. Maye gurbi guda ɗaya na tushe yayin kwafi shima yana iya yin illa.
Gyaran tarihi[gyara sashe | gyara masomin]
Eukaryotic DNA yana da hadadden tsari. Gabaɗaya an naɗe shi da sunadaran sunadaran da ake kira histones don samar da wani tsari da ake kira nucleosome . A nucleosome kunshi 2 sets na 4 tarihi: H2A, H2B, H3, da H4 . Bugu da ƙari, histone H1 yana ba da gudummawa ga tattarawar DNA a waje da nucleosome. Wasu gyare-gyaren enzymes na histone na iya ƙarawa ko cire ƙungiyoyi masu aiki zuwa histones, kuma waɗannan gyare-gyare suna tasiri matakin rubutun kwayoyin halitta da aka nannade a kusa da waɗancan tarihin da matakin kwafin DNA. Bayanan bayanan gyare-gyare na tarihi na ƙwayoyin lafiya da masu ciwon daji sun bambanta.
Idan aka kwatanta da ƙwayoyin lafiya, ƙwayoyin daji suna nuna raguwar nau'ikan histone H4 da monoacetylated (raguwar H4ac da H4me3). Bugu da ƙari, ƙirar linzamin kwamfuta sun nuna cewa raguwa a cikin histone H4R3 asymmetric dimethylation (H4R3me2a) na p19ARF mai gabatarwa yana da alaƙa tare da ƙarin ci gaba na ciwon daji da metastasis.[14] A cikin ƙirar linzamin kwamfuta, asarar histone H4 acetylation da trimethylation yana ƙaruwa yayin da ci gaban ƙwayar cuta ke ci gaba. Asarar histone H4 Lysine 16 acetylation ( H4K16ac ), wanda shine alamar tsufa a telomeres, musamman ya rasa acetylation. Wasu masana kimiyya suna fatan wannan asarar ta musamman na histone acetylation na iya yin yaƙi tare da mai hana histone deacetylase (HDAC) na musamman don SIRT1, HDAC na musamman don H4K16.
Sauran alamomin tarihin da ke hade da tumorigenesis sun hada da ƙarar deacetylation (raguwar acetylation) na histones H3 da H4, rage trimethylation na histone H3 Lysine 4 ( H3K4me3 ), da kuma ƙara monomethylation na histone H3 Lysine 9 ( H3K9me ) da kuma trimethylation na histone H3 Lysine 27me H3 Lysine 27 ( H3 . ). shi ne Waɗannan gyare-gyaren tarihi na iya yin shiru da ƙwayoyin cuta masu hana ƙari duk da raguwar methylation na tsibirin CpG na kwayoyin halitta (wani al'amari da yakan kunna kwayoyin halitta).
Wasu bincike sun mayar da hankali kan toshe aikin BRD4 a kan acetylated histones, wanda aka nuna don ƙara yawan bayyanar da furotin Myc, wanda ke da alaka da ciwon daji da yawa. Tsarin ci gaba na miyagun ƙwayoyi don ɗaure zuwa BRD4 abin lura ne ga haɗin gwiwa, buɗe hanyar da ƙungiyar ke ɗauka. [15]
Mai hana ƙwayar ƙwayar ƙwayar ƙwayar cuta ta p53 tana daidaita gyaran DNA kuma yana iya haifar da apoptosis a cikin ƙwayoyin da aka lalata. E Soto-Reyes da F Recillas-Targa sun bayyana mahimmancin furotin CTCF a cikin daidaita maganganun p53. CTCF, ko CCCTC daure factor, shine furotin yatsa na zinc wanda ke hana mai tallata p53 daga tara alamun tarihin danniya. A cikin wasu nau'ikan kwayoyin cutar kansa, furotin CTCF ba ya ɗaure kullum, kuma mai haɓaka p53 yana tara alamomin tarihi masu tsauri, yana haifar da bayyanar p53 don ragewa.
Maye gurbi a cikin na'urar epigenetic kanta na iya faruwa kuma, mai yuwuwar alhakin canza bayanan martaba na sel masu ciwon daji. Bambance-bambancen tarihi na dangin H2A ana kiyaye su sosai a cikin dabbobi masu shayarwa, suna taka muhimmiyar rawa wajen daidaita yawancin hanyoyin nukiliya ta hanyar canza tsarin chromatin . Ɗayan maɓalli na H2A, H2A. X, yana nuna lalacewar DNA, yana sauƙaƙe ɗaukar sunadaran gyaran DNA don maido da amincin genomic. Wani bambance-bambancen, H2A. Z, yana taka muhimmiyar rawa a duka kunna kwayoyin halitta da danniya. Babban darajar H2A. Ana gano furcin Z a cikin cututtukan daji da yawa kuma yana da alaƙa da haɓakar salon salula da rashin kwanciyar hankali.[16] Bambancin macroH2A1 na tarihi yana da mahimmanci a cikin cututtukan cututtukan daji da yawa, misali a cikin ciwon hanta.[17] Sauran hanyoyin sun haɗa da raguwa a cikin H4K16ac na iya haifar da ko dai raguwar ayyukan histone acetyltransferases (HATs) ko karuwa a deacetylation ta SIRT1. Hakazalika, maye gurbin frameshift wanda ba ya aiki a cikin HDAC2, histone deacetylase wanda ke aiki akan yawancin lysines na histone-tail, an hade shi da ciwon daji da ke nuna sauye-sauyen tsarin acetylation na histone. Wadannan binciken suna nuna wani tsari mai ban sha'awa don canza bayanan martaba na epigenetic ta hanyar hana enzymatic ko haɓakawa.
DNA lalacewa, lalacewa ta hanyar UV haske, ionizing radiation, muhalli guba, da kuma na rayuwa sinadarai, kuma zai iya haifar da genomic rashin zaman lafiya da kuma ciwon daji. Amsar lalacewa ta DNA zuwa ga karyawar DNA mai madauri biyu (DSB) ana yin sulhu a sashi ta gyare-gyaren tarihi. A DSB, MRE11 - RAD50 - NBS1 (MRN) hadaddun sunadaran suna ɗaukar ataxia telangiectasia mutated (ATM) kinase wanda phosphorylates Serine 129 na Histone 2A. MDC1, matsakanci na wurin binciken lalacewar DNA 1, yana ɗaure ga phosphopeptide, da phosphorylation na H2AX na iya yaduwa ta hanyar madaidaicin ra'ayi na daukar ma'aikata na MRN-ATM da phosphorylation. TIP60 acetylates da γH2AX, wanda aka sanya polyubiquitylated . RAP80, wani ɓangare na DNA na gyaran ƙwayar nono nau'in ciwon nono na nau'in 1 mai saukin kamuwa da furotin ( BRCA1 -A), yana ɗaure ubiquitin da aka haɗe zuwa histones. BRCA1-Aiki yana kama da sake zagayowar tantanin halitta a wurin bincike na G2/M, ba da damar lokaci don gyara DNA, ko apoptosis na iya farawa.
MicroRNA gene yin shiru[gyara sashe | gyara masomin]
A cikin dabbobi masu shayarwa, microRNAs (miRNAs), ya kasan ce suna daidaita kusan kashi 60% na ayyukan rufaffiyar kwayoyin halitta masu rufaffen furotin. [18] Wasu miRNA kuma suna yin shuru mai alaƙa da methylation a cikin ƙwayoyin kansa. Let-7 da miR15/16 suna taka muhimmiyar rawa wajen daidaita RAS da BCL2 oncogenes, kuma shirunsu yana faruwa a cikin ƙwayoyin kansa. Rage magana na miR-125b1, miRNA mai aiki azaman mai hana ƙari, an lura dashi a cikin prostate, ovarian, nono da glial cell cancers. Gwaje-gwajen in vitro sun nuna cewa miR-125b1 yana kaiwa ga kwayoyin halitta guda biyu, HER2/neu da ESR1, waɗanda ke da alaƙa da ciwon nono. DNA methylation, musamman hypermethylation, yana ɗaya daga cikin manyan hanyoyin da miR-125b1 aka rufe shi ta asali. A cikin marasa lafiya da ciwon nono, an lura da hypermethylation na tsibiran CpG da ke kusa da wurin fara rubutun. Asarar ɗaurin CTCF da haɓaka alamomin tarihi masu dannewa, H3K9me3 da H3K27me3, sun yi daidai da DNA methylation da shiru na miR-125b1. Ta hanyar injiniyanci, CTCF na iya aiki azaman yanki mai iyaka don dakatar da yaduwar DNA methylation. Sakamako daga gwaje-gwajen da Soto-Reyes et al. yana kuma nuna mummunan sakamako na methylation akan aiki da magana na miR-125b1. Saboda haka, sun yanke shawarar cewa DNA methylation yana da wani ɓangare na yin shiru da kwayar halitta. Bugu da ƙari, wasu miRNA's an rufe su da wuri da wuri a cikin ciwon nono, sabili da haka waɗannan miRNA na iya zama da amfani azaman alamun ƙari. kuma Yin shiru na epigenetic na kwayoyin miRNA ta hanyar DNA methylation aberrant wani lamari ne akai-akai a cikin kwayoyin cutar kansa; Kusan kashi ɗaya bisa uku na masu tallata miRNA masu aiki a cikin ƙwayoyin mammary na yau da kullun an sami hypermethylated a cikin ƙwayoyin kansar nono - wannan shine ninki da yawa fiye da yadda aka saba gani akan ƙwayoyin furotin.[19]
Metabolic recoding na epigenetics a cikin ciwon daji[gyara sashe | gyara masomin]
Dysregulation na metabolism yana ba da damar ƙwayoyin tumo don samar da tubalan ginin da ake buƙata da kuma daidaita alamun epigenetic don tallafawa farawa da ci gaba da ciwon daji. Canje-canje na rayuwa da ke haifar da ciwon daji yana canza yanayin yanayin epigenetic, musamman gyare-gyare akan histones da DNA, don haka inganta canji mara kyau, daidaitawa ga rashin isasshen abinci mai gina jiki, da metastasis. Tarin wasu metabolites a cikin ciwon daji na iya kai hari ga enzymes na epigenetic don canza yanayin yanayin epigenetic a duniya. Canje-canje na rayuwa da ke da alaƙa da ciwon daji yana haifar da ƙayyadaddun ƙayyadaddun wuraren sake canza alamun epigenetic. Ciwon daji epigenetics za a iya daidaita daidai da salon salula metabolism ta hanyar 1) kashi-amsa modulation na ciwon daji epigenetics ta metabolites; 2) takamaiman takamaiman ɗaukar aikin enzymes na rayuwa; da 3) niyya na Epigenetic enzymes ta hanyar siginar abinci mai gina jiki.[20]
MicroRNA da gyaran DNA[gyara sashe | gyara masomin]
Lalacewar DNA ya bayyana shine babban dalilin cutar kansa.[21][22] Idan gyaran DNA ya yi karanci, lalacewar DNA tana son taruwa. Irin wannan ɓarna na DNA da yawa na iya ƙara kurakuran maye gurbi yayin kwafin DNA saboda haɗakar fassarar kuskure. Lalacewar DNA ta wuce gona da iri na iya ƙara sauye-sauyen epigenetic saboda kurakurai yayin gyaran DNA.[23][24] Irin wannan maye gurbi da sauye-sauye na epigenetic na iya haifar da ciwon daji (duba m neoplasms ).
Canje-canjen layin ƙwayoyin ƙwayar cuta a cikin kwayoyin halittar DNA na gyarawa yana haifar da kashi 2-5 cikin ɗari kawai na cututtukan ciwon hanji . Koyaya, canza yanayin magana na microRNAs, yana haifar da raunin gyaran DNA, akai-akai ana danganta su da ciwon daji kuma yana iya zama muhimmiyar sanadin cutar kansa.
Yin wuce gona da iri na wasu miRNA na iya rage bayyana takamaiman sunadaran gyaran DNA kai tsaye. Wan et al.[25] ana magana da kwayoyin halittar DNA guda 6 waɗanda miRNA suka yi niyya kai tsaye a cikin baka: ATM (miR-421), RAD52 (miR-210, miR-373), RAD23B (miR-373), MSH2 (miR-21). ), BRCA1 (miR-182) da P53 (miR-504, miR-125b). Kwanan nan, Tessitore et al. [26] ya jera ƙarin ƙwayoyin gyaran DNA waɗanda ƙarin miRNAs ke niyya kai tsaye, gami da ATM (miR-18a, miR-101), DNA-PK (miR-101), ATR (miR-185), Wip1 (miR-16), MLH1, MSH2 da MSH6 (miR-155), ERCC3 da ERCC4 (miR-192) da UNG2 (mir-16, miR-34c da miR-199a). Daga cikin waɗannan miRNAs, miR-16, miR-18a, miR-21, miR-34c, miR-125b, miR-101, miR-155, miR-182, miR-185 da miR-192 suna cikin waɗanda Schnekenburger ya gano da kuma Diederich[27] kamar yadda aka yi yawa a cikin ciwon daji ta hanji ta hanyar epigenetic hypomethylation. Bayan bayyanar kowane ɗayan waɗannan miRNAs na iya haifar da raguwar bayyanar kwayar halittar DNA ɗin da ta ke niyya.
Har zuwa 15% na ƙarancin MLH1 -rauni a cikin cututtukan daji na hanji na lokaci-lokaci ya bayyana saboda wuce gona da iri na microRNA miR-155, wanda ke danne furcin MLH1. [28] Duk da haka, yawancin cututtukan daji na hanji 68 da aka rage tare da rage bayanin furotin MLH1 na rashin daidaituwa na DNA an gano cewa sun yi kasala saboda methylation na epigenetic na tsibirin CpG na MLH1 . [29]
A cikin 28% na glioblastomas, furotin gyaran DNA na MGMT ya gaza amma mai tallata MGMT ba methylated bane. [30] A cikin glioblastomas ba tare da methylated MGMT masu tallata ba, matakin microRNA miR-181d yana da alaƙa da alaƙa da bayanin furotin na MGMT kuma makasudin miR-181d kai tsaye shine MGMT mRNA 3'UTR ( yankin MGMT mRNA mafi girma uku da ba a fassara shi ba ). [30] Don haka, a cikin kashi 28% na glioblastomas, ƙara yawan magana na miR-181d da rage maganganun gyaran enzyme MGMT na iya zama sanadi. A cikin 29-66%[31][32] na glioblastomas, gyaran DNA yana da kasawa saboda epigenetic methylation na kwayar <i id="mwAUs">MGMT</i>, wanda ya rage bayanin furotin na MGMT.
Babban ƙungiyoyin motsi A ( HMGA ), sunadaran sunadaran AT-ƙugiya, ƙanana ne, nonhistone, sunadarai masu alaƙa da chromatin waɗanda zasu iya canza rubutun. MicroRNAs suna sarrafa bayanin sunadaran HMGA, kuma waɗannan sunadaran ( HMGA1 da HMGA2 ) abubuwa ne masu sarrafa rubutun chromatin na gine-gine. Palmieri et al. ya nuna cewa, a cikin kyallen takarda na al'ada, kwayoyin HGMA1 da HMGA2 an yi niyya (kuma saboda haka an rage su sosai a cikin magana) ta miR-15, miR-16, miR-26a, miR-196a2 da Let-7a .
Maganar HMGA kusan ba a iya gano shi a cikin bambance-bambancen kyallen jikin manya amma yana da girma a cikin cututtukan daji da yawa. Sunadaran HGMA polypeptides ne na ~ 100 ragowar amino acid wanda ke da tsarin tsari na zamani. Waɗannan sunadaran suna da yankuna uku masu inganci masu inganci, waɗanda ake kira AT hooks, waɗanda ke ɗaure ƙaramin tsagi na DNA mai arzikin AT a cikin takamaiman yankuna na DNA. Neoplasias na ɗan adam, ciki har da thyroid, prostatic, mahaifa, colorectal, pancreatic da ovarian carcinoma, suna nuna karuwa mai karfi na HMGA1a da HMGA1b sunadaran. [33] Mice masu canzawa tare da HMGA1 da aka yi niyya ga ƙwayoyin lymphoid suna haɓaka lymphoma mai ƙarfi, yana nuna cewa babban HMGA1 magana ba kawai yana da alaƙa da ciwon daji ba, amma cewa HMGA1 na iya aiki azaman oncogene don haifar da ciwon daji. [34] Baldassarre et al.,[35] ya nuna cewa sunadaran HMGA1 yana ɗaure zuwa yankin mai talla na DNA gyara gene BRCA1 kuma yana hana ayyukan mai gabatarwa BRCA1 . Sun kuma nuna cewa yayin da kawai 11% na ciwace-ciwacen nono ke da hypermethylation na BRCA1 gene, 82% na ciwon nono mai tsanani yana da ƙananan furotin na BRCA1, kuma yawancin waɗannan raguwa sun kasance saboda gyaran chromatin ta hanyar manyan matakan furotin HMGA1.
HMGA2 furotin na musamman yana kai hari ga mai tallata ERCC1, don haka rage bayyanar da wannan kwayar cutar ta DNA.[36] Maganganun furotin na ERCC1 ya gaza a cikin 100% na 47 da aka kimanta ciwon daji na hanji (ko da yake ba a san iyakar abin da HGMA2 ke ciki ba).[37]
Palmieri et al.[38] ya nuna cewa kowanne daga cikin miRNAs da ke hari kwayoyin HMGA sun ragu sosai a kusan dukkanin adenoma pituitary ɗan adam da aka yi nazari, idan aka kwatanta da glandan pituitary na yau da kullun. Daidai da ƙaƙƙarfan ƙa'ida na waɗannan miRNAs masu niyya HMGA, an sami haɓaka a cikin HMGA1 da takamaiman mRNAs na HMGA2. Uku daga cikin waɗannan microRNAs (miR-16, miR-196a da Let-7a))[39][40] suna da masu tallata methylated don haka ƙarancin magana a cikin ciwon hanji. Na biyu daga cikin waɗannan, miR-15 da miR-16, yankuna masu coding an rufe su ta asali cikin cutar kansa saboda ayyukan histone deacetylase . [41] Lokacin da aka bayyana waɗannan microRNAs a ƙaramin matakin, to ana bayyana sunadaran HMGA1 da HMGA2 a babban matakin. HMGA1 da HMGA2 manufa (rage maganganun) BRCA1 da ERCC1 DNA gyarawa. Don haka ana iya rage gyaran DNA, mai yiwuwa yana ba da gudummawa ga ci gaban kansa. [42]
Hanyoyin gyaran DNA[gyara sashe | gyara masomin]
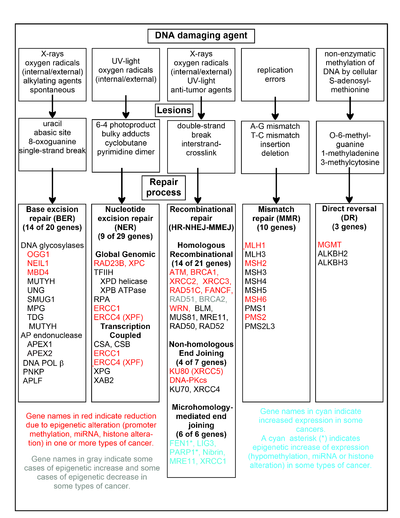
Jadawalin da ke cikin wannan sashe yana nuna wasu abubuwa masu lalata DNA akai-akai, misalan raunukan DNA da suke haifarwa, da hanyoyin da ke magance waɗannan lalacewar DNA. Aƙalla enzymes 169 ana amfani da su kai tsaye a cikin gyaran DNA ko kuma tasiri hanyoyin gyaran DNA. [43] Daga cikin waɗannan, 83 suna aiki kai tsaye wajen gyara nau'ikan lalacewar DNA guda 5 da aka kwatanta a cikin ginshiƙi.
Wasu daga cikin mafi kyawun binciken kwayoyin halitta na tsakiyar waɗannan hanyoyin gyara ana nuna su a cikin ginshiƙi. Zane-zanen jinsin da aka nuna a cikin ja, launin toka ko cyan suna nuna kwayoyin halitta akai-akai suna canzawa a cikin nau'ikan cututtukan daji daban-daban. Rubutun Wikipedia akan kowane ɗayan kwayoyin halitta da ja, launin toka ko cyan suka haskaka sun bayyana canjin (s) epigenetic da ciwon daji (s) waɗanda aka samo waɗannan abubuwan. Manyan labaran binciken gwaji guda biyu[44][45] kuma sun rubuta mafi yawan waɗannan raunin gyaran DNA na epigenetic a cikin cututtukan daji.
Kwayoyin halitta masu haske suna raguwa ko yin shiru akai-akai ta hanyoyin epigenetic a cikin cututtuka daban-daban. Lokacin da waɗannan kwayoyin halitta suna da ƙarancin magana ko rashi, lalacewar DNA na iya tarawa. Kurakuran maimaitawa da suka wuce waɗannan lalacewa (duba fassarar fassarar ) na iya haifar da haɓakar maye gurbi da, a ƙarshe, ciwon daji. Danniya na Epigenetic na kwayoyin gyaran DNA a cikin ingantattun hanyoyin gyaran DNA yana bayyana a matsayin tsakiya ga carcinogenesis .
Ƙwayoyin halitta guda biyu masu launin toka RAD51 da BRCA2, ana buƙatar su don gyara haɗin haɗin gwiwa . Wani lokaci ana nuna su a cikin epigenetically fiye da bayyanawa kuma wani lokacin ba a bayyana su a wasu cututtukan daji. Kamar yadda aka nuna a cikin labaran Wikipedia akan RAD51 da BRCA2, irin wannan ciwon daji yawanci suna da ƙarancin epigenetic a cikin wasu kwayoyin gyara DNA. Waɗannan ƙarancin gyare-gyare na iya haifar da ƙarin lalacewar DNA da ba a gyara su ba. Bayanin sama-sama na RAD51 da BRCA2 da aka gani a cikin waɗannan cututtukan na iya nuna matsi na zaɓi don ramuwa RAD51 ko BRCA2 fiye da bayyanawa da haɓaka gyare-gyare na haɗin gwiwa don aƙalla wani ɓangare na magance irin wannan lalacewar DNA. A waɗancan lokuta inda RAD51 ko BRCA2 ba a bayyana su ba, wannan da kansa zai haifar da ƙarin lalacewar DNA da ba a gyara ba. Kurakuran maimaitawa da suka wuce waɗannan lalacewa (duba fassarar fassarar ) na iya haifar da haɓakar maye gurbi da ciwon daji, ta yadda rashin bayyanar RAD51 ko BRCA2 zai zama carcinogenic a cikin kanta.
gyare-gyaren hutun madauri biyu, homology na 5-25 madaidaitan tushe guda 5-25 tsakanin maɗaurin biyun ya wadatar don daidaita madauri, amma ƙarshen da bai dace ba (flaps) yawanci yana nan. MMEJ yana cire ƙarin nucleotides (flaps) inda aka haɗa igiyoyi, sa'an nan kuma ya haɗa igiyoyin don ƙirƙirar helix na DNA marar kyau. MMEJ kusan ko da yaushe ya ƙunshi aƙalla ƙaramin gogewa, ta yadda ta zama hanyar mutagenic. [46] Fen1, Flap Sponglease a Mmej, shi ne epicinetally ya karu ta hanyar incarter hysomethyhybyhy ne kuma ya kare a cikin mafi cutar kansa na nono, ,[47][48][49] neuroblastomomas, [50] [51], [52] da huhu. [53] Har ila yau, PARP1 an fi bayyana shi sosai lokacin da yankin ETS mai tallata shi ya kasance hypomethylated na epigenetically, kuma wannan yana ba da gudummawa ga ci gaba zuwa ciwon daji na endometrial, [54] BRCA-mutated ovarian cancer, [55] da BRCA-mutated serous ovarian cancer. [56] Sauran kwayoyin halitta a cikin hanyar MMEJ suma an nuna su sosai a cikin adadin cutar kansa (duba MMEJ don taƙaitawa), kuma ana nuna su da shuɗi.
Matsakaicin ƙididdiga a cikin kwayoyin gyara DNA[gyara sashe | gyara masomin]
Rashin ƙarancin sunadaran gyaran DNA waɗanda ke aiki a cikin ingantattun hanyoyin gyaran DNA suna ƙara haɗarin maye gurbi. Adadin maye gurbi yana ƙaruwa sosai a cikin sel tare da maye gurbi a cikin gyare-gyaren rashin daidaituwa na DNA [57][58] ko a cikin gyaran haɗin gwiwa ( HRR ). [59] Mutanen da ke da maye gurbi a cikin kowane ɗayan 34 na gyaran DNA suna cikin haɗarin cutar kansa (duba lahani na gyaran DNA da haɓaka haɗarin kansa ).
A cikin cututtukan daji na lokaci-lokaci, ana samun rashi a gyaran DNA lokaci-lokaci saboda maye gurbi a cikin kwayar halittar DNA, amma yawanci ana ragewa ko rashin bayyanar kwayoyin halittar DNA saboda sauye-sauyen epigenetic da ke rage ko yin shiru. Alal misali, don ciwon daji na launi na 113 da aka yi nazari a cikin jerin, hudu kawai suna da maye gurbi a cikin DNA na gyaran MGMT, yayin da mafi yawan sun rage MGMT magana saboda methylation na yankin mai gabatarwa na MGMT (canjin epigenetic). [60] Hakazalika, daga cikin lokuta 119 na rashin daidaiton gyare-gyare-rashin ciwon daji na launi waɗanda ba su da gyaran DNA na PMS2 magana, sunadarin PMS2 ya gaza a cikin 6 saboda maye gurbi a cikin kwayoyin PMS2, yayin da a cikin lokuta 103 PMS2 magana ta kasa saboda an danne abokin tarayya MLH1. saboda mai gabatarwa methylation (protein PMS2 ba shi da kwanciyar hankali idan babu MLH1). A cikin sauran shari'o'in 10, asarar bayanin PMS2 ya kasance mai yuwuwa saboda haɓakar epigenetic na microRNA, miR-155, wanda ke daidaita MLH1.
Lalacewar Epigenetic a cikin kwayoyin halittar DNA na gyaran gyare-gyare sun kasance akai-akai a cikin ciwon daji. A cikin tebur, an ƙididdige ciwon daji da yawa don ragewa ko rashin bayyanar jigon gyaran DNA na sha'awa, kuma mitar da aka nuna shi ne yawan ciwon daji da ke da ƙarancin epigenetic na maganganun kwayoyin halitta. Irin wannan raunin epigenetic yana iya tasowa da wuri a cikin carcinogenesis, tun da yake ana samun su akai-akai (ko da yake a ɗan ƙananan mita) a cikin lahani da ke kewaye da ciwon daji wanda ciwon daji zai iya tasowa (duba Table).
| Cancer | Gene | Frequency in Cancer | Frequency in Field Defect | Ref. |
|---|---|---|---|---|
| Colorectal | MGMT | 46% | 34% | [61] |
| Colorectal | MGMT | 47% | 11% | [62] |
| Colorectal | MGMT | 70% | 60% | [63] |
| Colorectal | MSH2 | 13% | 5% | [62] |
| Colorectal | ERCC1 | 100% | 40% | [64] |
| Colorectal | PMS2 | 88% | 50% | [64] |
| Colorectal | XPF | 55% | 40% | [64] |
| Head and Neck | MGMT | 54% | 38% | [65] |
| Head and Neck | MLH1 | 33% | 25% | [66] |
| Head and Neck | MLH1 | 31% | 20% | [67] |
| Stomach | MGMT | 88% | 78% | [68] |
| Stomach | MLH1 | 73% | 20% | [69] |
| Esophagus | MLH1 | 77%–100% | 23%–79% | [70] |
Ya bayyana cewa ana iya farawa da kansa akai-akai ta hanyar raguwar epigenetic a cikin furci ɗaya ko fiye da gyaran enzymes na DNA. Rage gyaran DNA na iya ba da damar tara lalacewar DNA. Kuskuren haɗakar fassarar fassarar wasu daga cikin waɗannan lalacewar DNA na iya haifar da maye gurbi tare da zaɓin fa'ida. Faci na clonal tare da zaɓin zaɓi na iya girma kuma ya fita gasa sel makwabta, yana haifar da lahani na filin . Duk da yake babu wata fa'ida ta zahiri ga tantanin halitta don ya rage gyaran DNA, za a iya ɗaukar jigilar kwayar halittar DNA ɗin tare da fasinja lokacin da sel masu zaɓin maye gurbi. A cikin sel waɗanda ke ɗauke da ƙayyadaddun kwayoyin halittar DNA na gyaran DNA da maye gurbi tare da zaɓin zaɓi, ƙarin lalacewar DNA za su taru, kuma waɗannan na iya haifar da ƙarin maye gurbi tare da zaɓi mafi girma. Lalacewar Epigenetic a cikin gyaran DNA na iya ba da gudummawa ga sifa mai girman mitar maye gurbi a cikin kwayoyin cutar kansa, kuma yana haifar da ci gaban carcinogenic.
Ciwon daji suna da babban matakan rashin zaman lafiyar kwayoyin halitta, wanda ke da alaƙa da yawan maye gurbi . Yawaitar maye gurbi yana ƙara yuwuwar takamaiman maye gurbi da ke faruwa waɗanda ke kunna oncogenes da ƙwayoyin cuta masu hana ƙari, wanda ke haifar da carcinogenesis . Dangane da tsarin tsarin kwayoyin halitta gaba daya, ana samun ciwon daji suna da dubunnan zuwa daruruwan dubbai na maye gurbi a dukkan kwayoyin halittarsu.[71] (Haka kuma duba mitocin maye gurbi a cikin cututtukan daji . ) Idan aka kwatanta, mitar maye gurbi a cikin dukkanin kwayoyin halitta tsakanin tsararraki ga mutane (iyaye da yaro) kusan sabbin maye gurbi 70 ne a kowane zamani.[72][73] A cikin yankuna masu rikodin sunadaran kwayoyin halitta, akwai kusan 0.35 maye gurbi tsakanin zuriyar iyaye/yara (kasa da furotin da aka canza a kowane zamani). [74] Gabaɗayan jerin kwayoyin halitta a cikin ƙwayoyin jini don wasu tagwaye iri ɗaya masu shekaru 100 kawai sun sami bambance-bambancen somatic 8, kodayake ba za a iya gano bambancin somatic da ke faruwa a ƙasa da kashi 20% na ƙwayoyin jini ba. [75]
Yayin da lalacewar DNA na iya haifar da maye gurbi ta hanyar haɗin fassarar kuskure, lalacewar DNA kuma na iya haifar da sauye-sauye na epigenetic yayin tafiyar da DNA mara kyau. Lalacewar DNA da ke taruwa saboda lahani na gyaran DNA na epigenetic na iya zama tushen haɓakar sauye-sauyen epigenetic da aka samu a yawancin ƙwayoyin cuta a cikin cututtukan daji. A cikin binciken farko, duban ƙayyadaddun saiti na masu tallata rubutu, Fernandez et al. [76] yayi nazarin bayanan methylation DNA na ciwace-ciwacen farko na 855. Kwatanta kowane nau'in ƙwayar cuta tare da nama na yau da kullun na yau da kullun, rukunin tsibirin CpG 729 (55% na rukunin 1322 CpG da aka kimanta) sun nuna bambancin DNA methylation. Daga cikin waɗannan rukunin yanar gizon, 496 sun kasance hypermethylated (danne) kuma 233 sun kasance hypomethylated (an kunna). Don haka, akwai babban matakin haɓakar epigenetic mai haɓaka methylation a cikin ciwace-ciwace. Wasu daga cikin waɗannan sauye-sauye na epigenetic na iya taimakawa wajen ci gaban ciwon daji.
- ↑ Sharma, S.; Kelly, T. K.; Jones, P. A. (2010-01-01). "Epigenetics in cancer". Carcinogenesis (in Turanci). 31 (1): 27–36. doi:10.1093/carcin/bgp220. ISSN 0143-3334. PMC 2802667. PMID 19752007.
- ↑ Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW (March 2013). "Cancer genome landscapes". Science. 339 (6127): 1546–58. Bibcode:2013Sci...339.1546V. doi:10.1126/science.1235122. PMC 3749880. PMID 23539594.
- ↑ Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP (September 2010). "Orphan CpG islands identify numerous conserved promoters in the mammalian genome". PLOS Genetics. 6 (9): e1001134. doi:10.1371/journal.pgen.1001134. PMC 2944787. PMID 20885785.
- ↑ Wei J, Li G, Dang S, Zhou Y, Zeng K, Liu M (2016). "Discovery and Validation of Hypermethylated Markers for Colorectal Cancer". Disease Markers. 2016: 2192853. doi:10.1155/2016/2192853. PMC 4963574. PMID 27493446.
- ↑ Beggs AD, Jones A, El-Bahrawy M, El-Bahwary M, Abulafi M, Hodgson SV, Tomlinson IP (April 2013). "Whole-genome methylation analysis of benign and malignant colorectal tumours". The Journal of Pathology. 229 (5): 697–704. doi:10.1002/path.4132. PMC 3619233. PMID 23096130.
- ↑ Novak K (December 2004). "Epigenetics changes in cancer cells". MedGenMed. 6 (4): 17. PMC 1480584. PMID 15775844.
- ↑ Banno K, Kisu I, Yanokura M, Tsuji K, Masuda K, Ueki A, Kobayashi Y, Yamagami W, Nomura H, Tominaga E, Susumu N, Aoki D (September 2012). "Epimutation and cancer: a new carcinogenic mechanism of Lynch syndrome (Review)". International Journal of Oncology. 41 (3): 793–7. doi:10.3892/ijo.2012.1528. PMC 3582986. PMID 22735547.
- ↑ Bird A (January 2002). "DNA methylation patterns and epigenetic memory". Genes & Development. 16 (1): 6–21. doi:10.1101/gad.947102. PMID 11782440.
- ↑ Herman, James G.; Graff, Jeremy R.; Myöhänen, Sanna; Nelkin, Barry D.; Baylin, Stephen B. (September 1996). "Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands". Proceedings of the National Academy of Sciences of the United States of America. 93 (18): 9821–6. Bibcode:1996PNAS...93.9821H. doi:10.1073/pnas.93.18.9821. PMC 38513. PMID 8790415.
- ↑ Wong NC, Craig JM (2011). Epigenetics: A Reference Manual. Norfolk, England: Caister Academic Press. ISBN 978-1-904455-88-2.
- ↑ De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA (May 2012). "DNA methylation screening identifies driver epigenetic events of cancer cell survival". Cancer Cell. 21 (5): 655–67. doi:10.1016/j.ccr.2012.03.045. PMC 3395886. PMID 22624715.
- ↑ Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR (May 2010). "RNAi and heterochromatin repress centromeric meiotic recombination". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8701–5. Bibcode:2010PNAS..107.8701E. doi:10.1073/pnas.0914160107. PMC 2889303. PMID 20421495.
- ↑ Timp W, Feinberg AP (July 2013). "Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host". Nature Reviews. Cancer (in Turanci). 13 (7): 497–510. doi:10.1038/nrc3486. PMC 4636434. PMID 23760024.
- ↑ Aprelikova O, Chen K, El Touny LH, Brignatz-Guittard C, Han J, Qiu T, Yang HH, Lee MP, Zhu M, Green JE (Apr 2016). "The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis". Clinical Epigenetics. 8 (38): 38. doi:10.1186/s13148-016-0205-6. PMC 4831179. PMID 27081402.
- ↑ Maxmen A (August 2012). "Cancer research: Open ambition". Nature. 488 (7410): 148–50. Bibcode:2012Natur.488..148M. doi:10.1038/488148a. PMID 22874946.
- ↑ Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR (May 2010). "RNAi and heterochromatin repress centromeric meiotic recombination". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8701–5. Bibcode:2010PNAS..107.8701E. doi:10.1073/pnas.0914160107. PMC 2889303. PMID 20421495.
- ↑ Rappa F, Greco A, Podrini C, Cappello F, Foti M, Bourgoin L, Peyrou M, Marino A, Scibetta N, Williams R, Mazzoccoli G, Federici M, Pazienza V, Vinciguerra M (2013). Folli F (ed.). "Immunopositivity for histone macroH2A1 isoforms marks steatosis-associated hepatocellular carcinoma". PLOS ONE. 8 (1): e54458. Bibcode:2013PLoSO...854458R. doi:10.1371/journal.pone.0054458. PMC 3553099. PMID 23372727.
- ↑ Friedman RC, Farh KK, Burge CB, Bartel DP (January 2009). "Most mammalian mRNAs are conserved targets of microRNAs". Genome Research. 19 (1): 92–105. doi:10.1101/gr.082701.108. PMC 2612969. PMID 18955434.
- ↑ Vrba L, Muñoz-Rodríguez JL, Stampfer MR, Futscher BW (2013). "miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer". PLOS ONE. 8 (1): e54398. Bibcode:2013PLoSO...854398V. doi:10.1371/journal.pone.0054398. PMC 3547033. PMID 23342147.
- ↑ Wang YP, Lei QY (May 2018). "Metabolic recoding of epigenetics in cancer". Cancer Communications. 38 (1): 25. doi:10.1186/s40880-018-0302-3. PMC 5993135. PMID 29784032.
- ↑ Kastan MB (2008). "DNA damage responses: mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture". Mol. Cancer Res. 6 (4): 517–24. doi:10.1158/1541-7786.MCR-08-0020. PMID 18403632.
- ↑ Bernstein C, Prasad AR, Nfonsam V, Bernstein H (2013). "Chapter 16: DNA Damage, DNA Repair and Cancer". In Chen C (ed.). New Research Directions in DNA Repair. p. 413. ISBN 978-953-51-1114-6.
- ↑ O'Hagan HM, Mohammad HP, Baylin SB (August 2008). "Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island". PLOS Genetics. 4 (8): e1000155. doi:10.1371/journal.pgen.1000155. PMC 2491723. PMID 18704159.
- ↑ Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV (July 2007). "DNA damage, homology-directed repair, and DNA methylation". PLOS Genetics. 3 (7): e110. doi:10.1371/journal.pgen.0030110. PMC 1913100. PMID 17616978.
- ↑ Wan G, Mathur R, Hu X, Zhang X, Lu X (September 2011). "miRNA response to DNA damage". Trends in Biochemical Sciences. 36 (9): 478–84. doi:10.1016/j.tibs.2011.06.002. PMC 3532742. PMID 21741842.
- ↑ Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E (2014). "MicroRNAs in the DNA Damage/Repair Network and Cancer". International Journal of Genomics. 2014: 820248. doi:10.1155/2014/820248. PMC 3926391. PMID 24616890.
- ↑ Schnekenburger M, Diederich M (March 2012). "Epigenetics Offer New Horizons for Colorectal Cancer Prevention". Current Colorectal Cancer Reports. 8 (1): 66–81. doi:10.1007/s11888-011-0116-z. PMC 3277709. PMID 22389639.
- ↑ Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, Costinean S, Sandhu SK, Nuovo GJ, Alder H, Gafa R, Calore F, Ferracin M, Lanza G, Volinia S, Negrini M, McIlhatton MA, Amadori D, Fishel R, Croce CM (April 2010). "Modulation of mismatch repair and genomic stability by miR-155". Proceedings of the National Academy of Sciences of the United States of America. 107 (15): 6982–7. Bibcode:2010PNAS..107.6982V. doi:10.1073/pnas.1002472107. JSTOR 25665289. PMC 2872463. PMID 20351277.
- ↑ Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schär P, Jiricny J, Marra G (May 2005). "Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer". Gastroenterology. 128 (5): 1160–71. doi:10.1053/j.gastro.2005.01.056. PMID 15887099.
- ↑ 30.0 30.1 Empty citation (help)
- ↑ Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, Jiang T, Chen CC (June 2012). "miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression". Neuro-Oncology. 14 (6): 712–9. doi:10.1093/neuonc/nos089. PMC 3367855. PMID 22570426.
- ↑ Spiegl-Kreinecker S, Pirker C, Filipits M, Lötsch D, Buchroithner J, Pichler J, Silye R, Weis S, Micksche M, Fischer J, Berger W (January 2010). "O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients". Neuro-Oncology. 12 (1): 28–36. doi:10.1093/neuonc/nop003. PMC 2940563. PMID 20150365.
- ↑ Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V (September 2004). "Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer". FEBS Letters. 574 (1–3): 1–8. doi:10.1016/j.febslet.2004.08.013. PMID 15358530. S2CID 28903539.
- ↑ Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ, Huso DL, Resar LM (May 2004). "The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia". Cancer Research. 64 (10): 3371–5. doi:10.1158/0008-5472.CAN-04-0044. PMID 15150086.
- ↑ Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F, Fedele M, Pierantoni G, Croce CM, Fusco A (April 2003). "Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma". Molecular and Cellular Biology. 23 (7): 2225–38. doi:10.1128/MCB.23.7.2225-2238.2003. PMC 150734. PMID 12640109.
- ↑ Borrmann L, Schwanbeck R, Heyduk T, Seebeck B, Rogalla P, Bullerdiek J, Wisniewski JR (December 2003). "High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity". Nucleic Acids Research. 31 (23): 6841–51. doi:10.1093/nar/gkg884. PMC 290254. PMID 14627817.
- ↑ Facista A, Nguyen H, Lewis C, Prasad AR, Ramsey L, Zaitlin B, Nfonsam V, Krouse RS, Bernstein H, Payne CM, Stern S, Oatman N, Banerjee B, Bernstein C (April 2012). "Deficient expression of DNA repair enzymes in early progression to sporadic colon cancer". Genome Integrity. 3 (1): 3. doi:10.1186/2041-9414-3-3. PMC 3351028. PMID 22494821.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedPalmieri - ↑ Schnekenburger M, Diederich M (March 2012). "Epigenetics Offer New Horizons for Colorectal Cancer Prevention". Current Colorectal Cancer Reports. 8 (1): 66–81. doi:10.1007/s11888-011-0116-z. PMC 3277709. PMID 22389639.
- ↑ Malumbres M (2013). "miRNAs and cancer: an epigenetics view". Molecular Aspects of Medicine. 34 (4): 863–74. doi:10.1016/j.mam.2012.06.005. PMC 5791883. PMID 22771542.
- ↑ Sampath D, Liu C, Vasan K, Sulda M, Puduvalli VK, Wierda WG, Keating MJ (February 2012). "Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia". Blood. 119 (5): 1162–72. doi:10.1182/blood-2011-05-351510. PMC 3277352. PMID 22096249.
- ↑ Bernstein C, Prasad AR, Nfonsam V, Bernstein H (2013). "Chapter 16: DNA Damage, DNA Repair and Cancer". In Chen C (ed.). New Research Directions in DNA Repair. p. 413. ISBN 978-953-51-1114-6.
- ↑ Human DNA Repair Genes, 15 April 2014, MD Anderson Cancer Center, University of Texas
- ↑ Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM (February 2013). "MicroRNA-182-5p targets a network of genes involved in DNA repair". RNA. 19 (2): 230–42. doi:10.1261/rna.034926.112. PMC 3543090. PMID 23249749.
- ↑ Chaisaingmongkol J, Popanda O, Warta R, Dyckhoff G, Herpel E, Geiselhart L, Claus R, Lasitschka F, Campos B, Oakes CC, Bermejo JL, Herold-Mende C, Plass C, Schmezer P (December 2012). "Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma". Oncogene. 31 (49): 5108–16. doi:10.1038/onc.2011.660. PMID 22286769.
- ↑ Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA (September 2005). "Modulation of DNA end joining by nuclear proteins". The Journal of Biological Chemistry. 280 (36): 31442–9. doi:10.1074/jbc.M503776200. PMID 16012167.
- ↑ Krause A, Combaret V, Iacono I, Lacroix B, Compagnon C, Bergeron C, Valsesia-Wittmann S, Leissner P, Mougin B, Puisieux A (July 2005). "Genome-wide analysis of gene expression in neuroblastomas detected by mass screening" (PDF). Cancer Letters. 225 (1): 111–20. doi:10.1016/j.canlet.2004.10.035. PMID 15922863.
- ↑ Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Yoo HS, Kim YS, Kim JG, Kim NS (January 2005). "Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells". Clinical Cancer Research. 11 (2 Pt 1): 473–82. PMID 15701830.
- ↑ Wang K, Xie C, Chen D (May 2014). "Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis". International Journal of Molecular Medicine. 33 (5): 1268–74. doi:10.3892/ijmm.2014.1682. PMID 24590400.
- ↑ Li D, Bi FF, Cao JM, Cao C, Li CY, Liu B, Yang Q (January 2014). "Poly (ADP-ribose) polymerase 1 transcriptional regulation: a novel crosstalk between histone modification H3K9ac and ETS1 motif hypomethylation in BRCA1-mutated ovarian cancer". Oncotarget. 5 (1): 291–7. doi:10.18632/oncotarget.1549. PMC 3960209. PMID 24448423.
- ↑ Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M (April 2003). "Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays". The American Journal of Pathology. 162 (4): 1151–62. doi:10.1016/S0002-9440(10)63911-9. PMC 1851213. PMID 12651607.
- ↑ Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD (October 2003). "Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer". Oncogene. 22 (46): 7243–6. doi:10.1038/sj.onc.1206977. PMID 14562054.
- ↑ Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, Zeng G, Horvath S, Belldegrun AS (August 2006). "Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score". BJU International. 98 (2): 445–51. doi:10.1111/j.1464-410X.2006.06224.x. PMID 16879693. S2CID 22165252.
- ↑ Bi FF, Li D, Yang Q (February 2013). "Promoter hypomethylation, especially around the E26 transformation-specific motif, and increased expression of poly (ADP-ribose) polymerase 1 in BRCA-mutated serous ovarian cancer". BMC Cancer. 13: 90. doi:10.1186/1471-2407-13-90. PMC 3599366. PMID 23442605.
- ↑ Empty citation (help)
- ↑ Empty citation (help)
- ↑ Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM (April 1997). "Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2". Proceedings of the National Academy of Sciences of the United States of America. 94 (7): 3122–7. Bibcode:1997PNAS...94.3122N. doi:10.1073/pnas.94.7.3122. JSTOR 41786. PMC 20332. PMID 9096356.
- ↑ Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM (December 2006). "Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6". Carcinogenesis. 27 (12): 2402–8. doi:10.1093/carcin/bgl079. PMC 2612936. PMID 16728433.
- ↑ Empty citation (help)
- ↑ Halford S, Rowan A, Sawyer E, Talbot I, Tomlinson I (June 2005). "O(6)-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions". Gut. 54 (6): 797–802. doi:10.1136/gut.2004.059535. PMC 1774551. PMID 15888787.
- ↑ Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JP (September 2005). "MGMT promoter methylation and field defect in sporadic colorectal cancer". Journal of the National Cancer Institute. 97 (18): 1330–8. doi:10.1093/jnci/dji275. PMID 16174854.
- ↑ 62.0 62.1 Lee KH, Lee JS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Lee JH (October 2011). "Promoter methylation status of hMLH1, hMSH2, and MGMT genes in colorectal cancer associated with adenoma-carcinoma sequence". Langenbeck's Archives of Surgery. 396 (7): 1017–26. doi:10.1007/s00423-011-0812-9. PMID 21706233. S2CID 8069716.
- ↑ Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, Seruca R, Gaub MP, Legrain M, Collura A, Lascols O, Tiret E, Fléjou JF, Duval A (November 2010). "Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers". Gut. 59 (11): 1516–26. doi:10.1136/gut.2009.194787. PMID 20947886. S2CID 206950452.
- ↑ 64.0 64.1 64.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedFacista - ↑ Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer-Dubowska W (February 2011). "Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa". Oral Oncology. 47 (2): 104–7. doi:10.1016/j.oraloncology.2010.11.006. PMID 21147548.
- ↑ Zuo C, Zhang H, Spencer HJ, Vural E, Suen JY, Schichman SA, Smoller BR, Kokoska MS, Fan CY (October 2009). "Increased microsatellite instability and epigenetic inactivation of the hMLH1 gene in head and neck squamous cell carcinoma". Otolaryngology–Head and Neck Surgery. 141 (4): 484–90. doi:10.1016/j.otohns.2009.07.007. PMID 19786217. S2CID 8357370.
- ↑ Tawfik HM, El-Maqsoud NM, Hak BH, El-Sherbiny YM (2011). "Head and neck squamous cell carcinoma: mismatch repair immunohistochemistry and promoter hypermethylation of hMLH1 gene". American Journal of Otolaryngology. 32 (6): 528–36. doi:10.1016/j.amjoto.2010.11.005. PMID 21353335.
- ↑ Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ (November 2009). "Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions". Human Pathology. 40 (11): 1534–42. doi:10.1016/j.humpath.2009.01.029. PMID 19695681.
- ↑ Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G, Wani R, Wani K (2012). "Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley". Asian Pacific Journal of Cancer Prevention. 13 (8): 4177–81. doi:10.7314/APJCP.2012.13.8.4177. PMID 23098428.
- ↑ Agarwal A, Polineni R, Hussein Z, Vigoda I, Bhagat TD, Bhattacharyya S, Maitra A, Verma A (2012). "Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma". International Journal of Clinical and Experimental Pathology. 5 (5): 382–96. PMC 3396065. PMID 22808291.
- ↑ Tuna M, Amos CI (November 2013). "Genomic sequencing in cancer". Cancer Letters. 340 (2): 161–70. doi:10.1016/j.canlet.2012.11.004. PMC 3622788. PMID 23178448.
- ↑ Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ (April 2010). "Analysis of genetic inheritance in a family quartet by whole-genome sequencing". Science. 328 (5978): 636–9. Bibcode:2010Sci...328..636R. doi:10.1126/science.1186802. PMC 3037280. PMID 20220176.
- ↑ Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, Vives L, O'Roak BJ, Sudmant PH, Shendure J, Abney M, Ober C, Eichler EE (November 2012). "Estimating the human mutation rate using autozygosity in a founder population". Nature Genetics. 44 (11): 1277–81. doi:10.1038/ng.2418. PMC 3483378. PMID 23001126.
- ↑ Keightley PD (February 2012). "Rates and fitness consequences of new mutations in humans". Genetics. 190 (2): 295–304. doi:10.1534/genetics.111.134668. PMC 3276617. PMID 22345605.
- ↑ Ye K, Beekman M, Lameijer EW, Zhang Y, Moed MH, van den Akker EB, Deelen J, Houwing-Duistermaat JJ, Kremer D, Anvar SY, Laros JF, Jones D, Raine K, Blackburne B, Potluri S, Long Q, Guryev V, van der Breggen R, Westendorp RG, 't Hoen PA, den Dunnen J, van Ommen GJ, Willemsen G, Pitts SJ, Cox DR, Ning Z, Boomsma DI, Slagboom PE (December 2013). "Aging as accelerated accumulation of somatic variants: whole-genome sequencing of centenarian and middle-aged monozygotic twin pairs" (PDF). Twin Research and Human Genetics. 16 (6): 1026–32. doi:10.1017/thg.2013.73. PMID 24182360.
- ↑ Empty citation (help)
